event
Third (Virtual) Workshop of the HPDaSc Project

Event Info
-
Date
10-12-2020
-
Location
Online
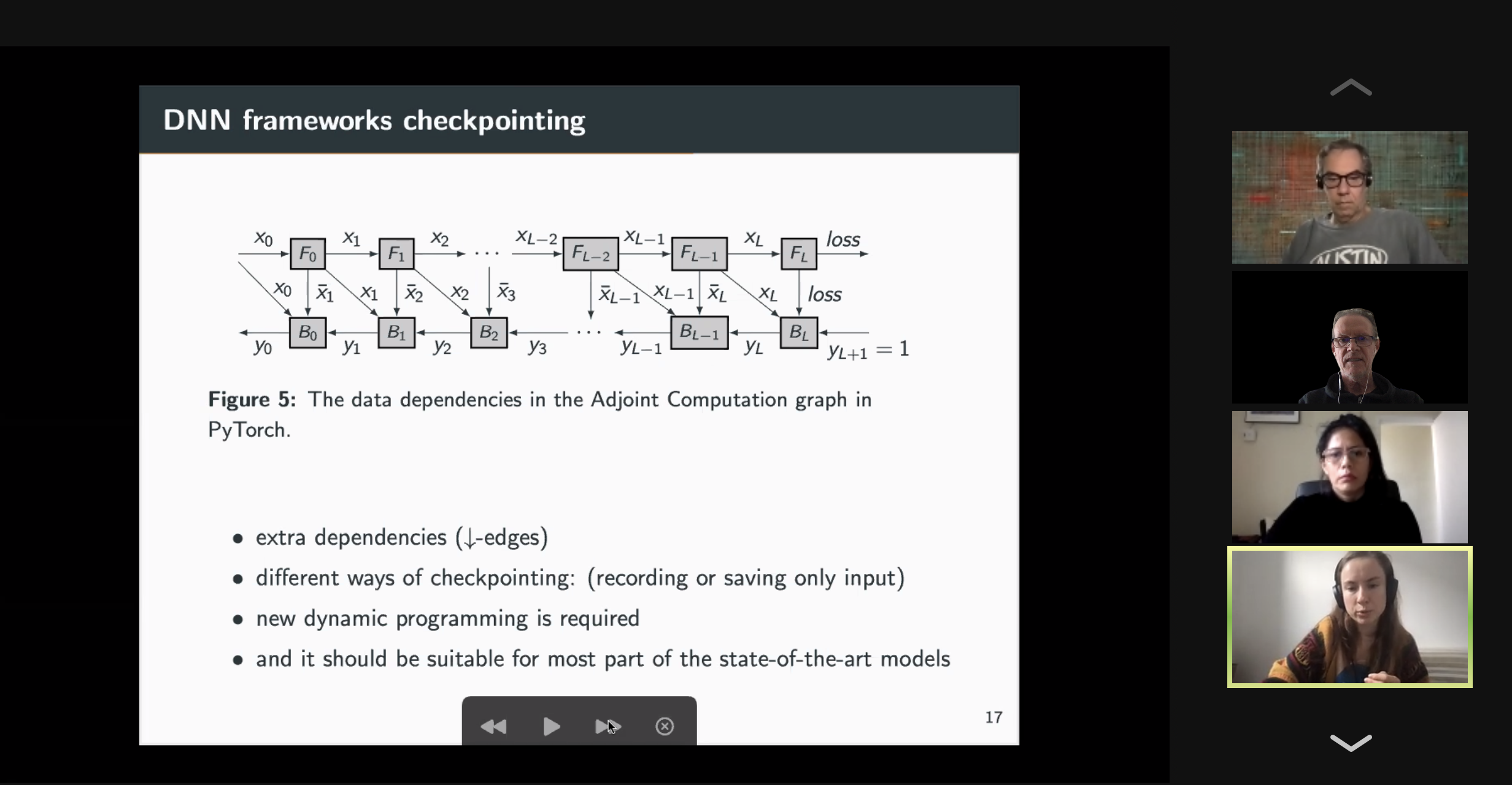
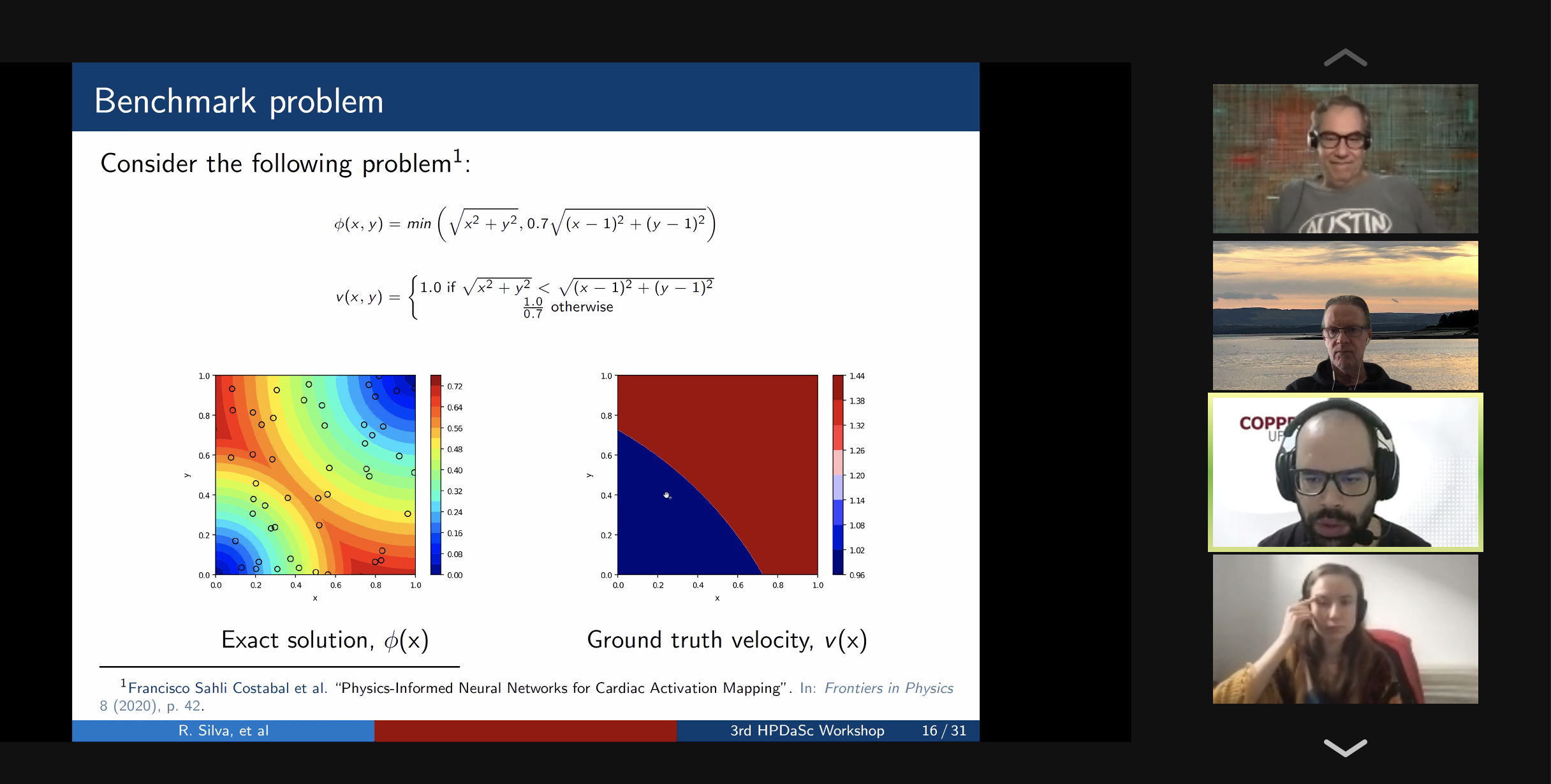
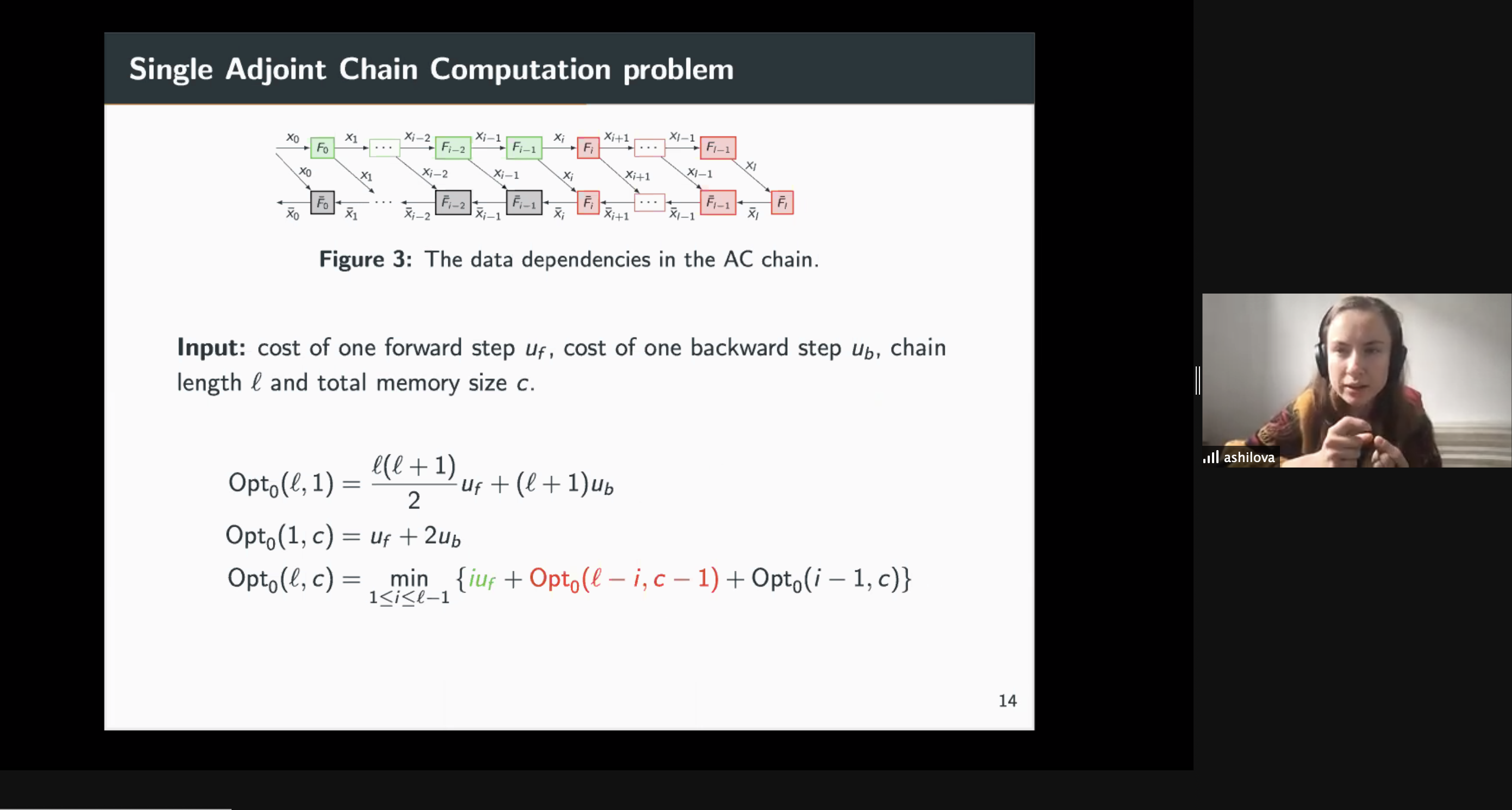
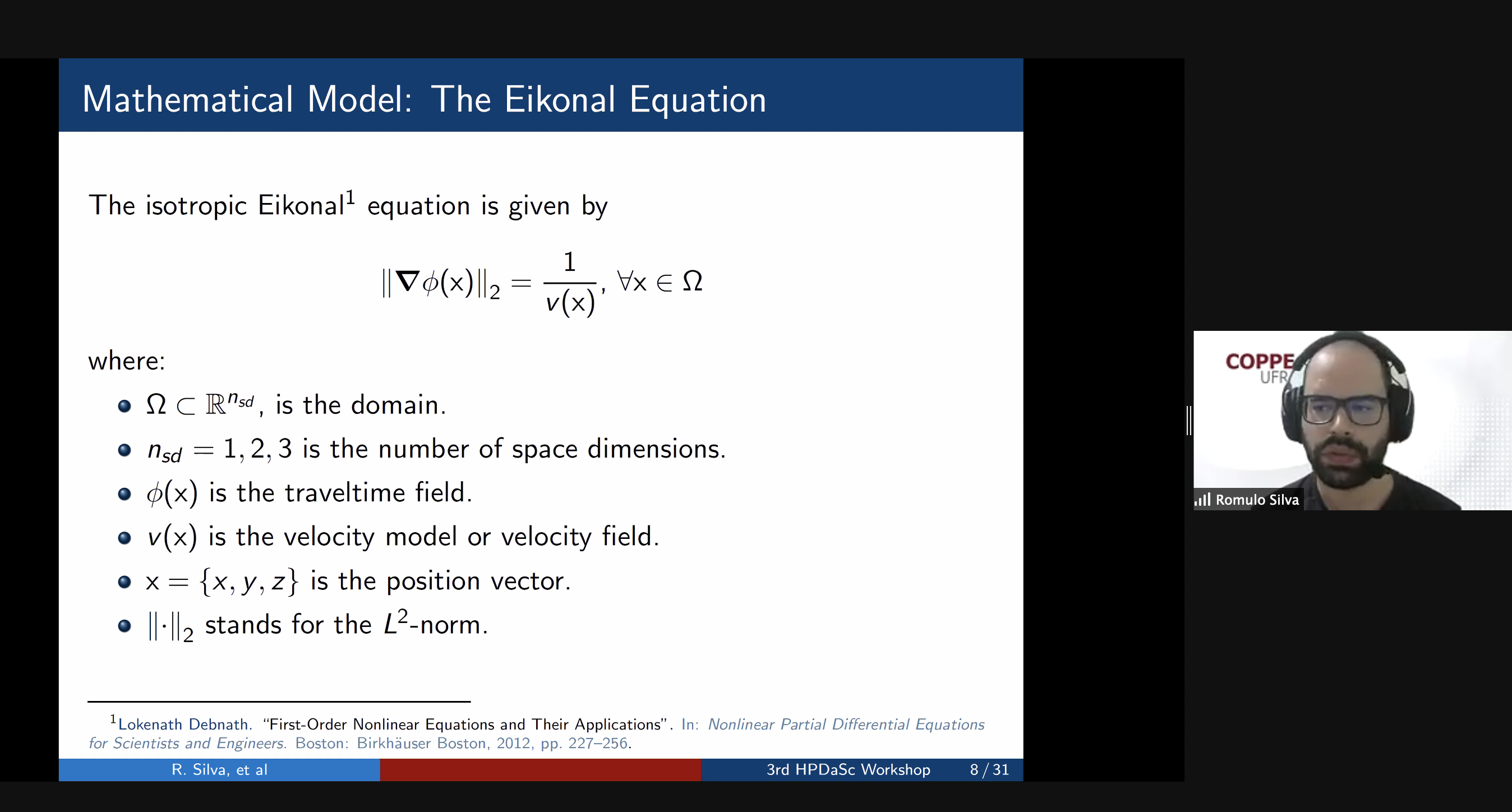
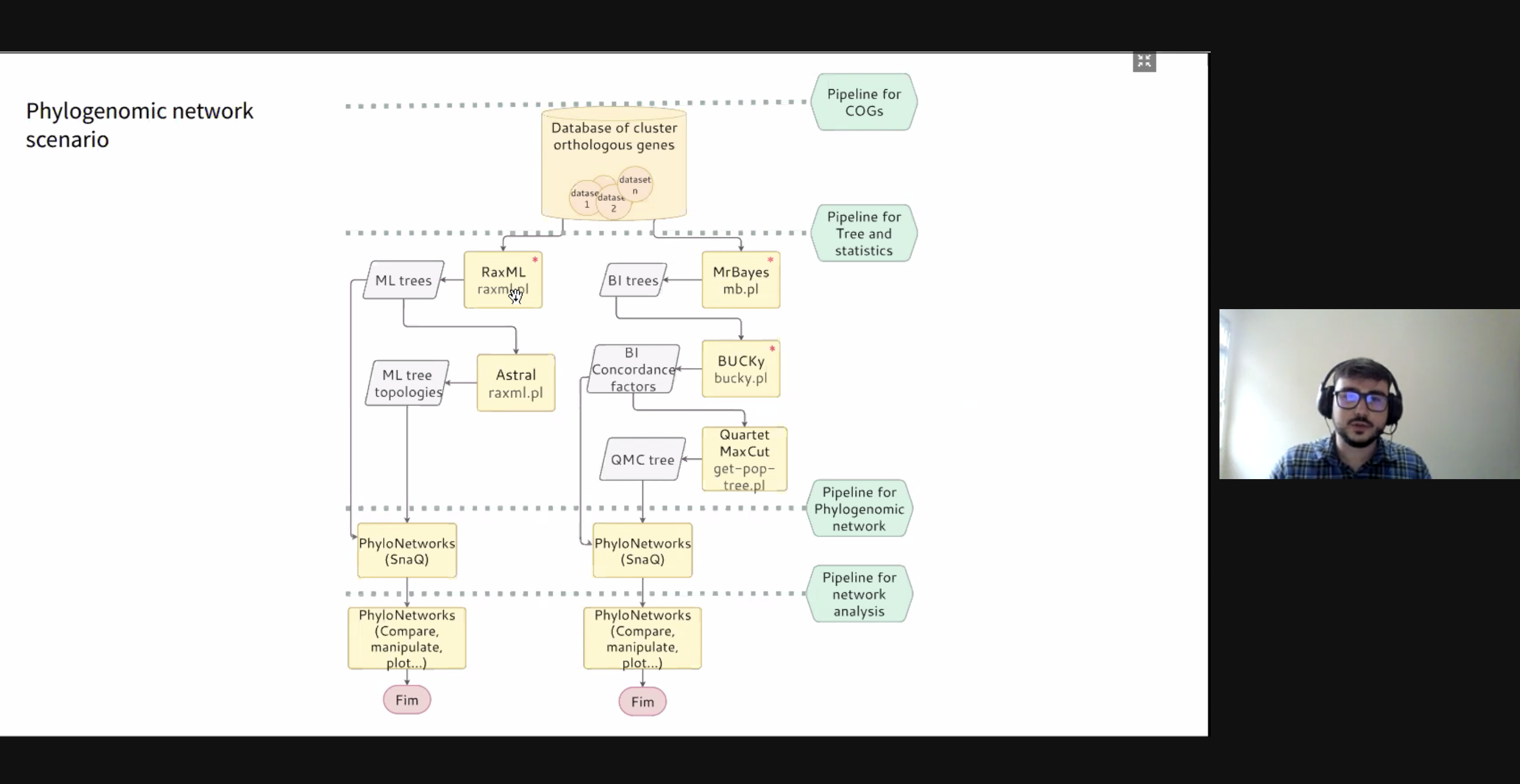
10:00 – 10:30 / 14:00 – 14:30 Title: Optimal Re-Materialization Strategies for Heterogeneous Chains: How to Train Deep Neural Networks with Limited Memory, Alena Shilova, Olivier Beaumont, Lionel Eyraud-Dubois, Julien Herrmann, Alexis Joly Abstract: We introduce a new activation re-materialization strategy, which allows significantly decreasing memory usage when training Deep Neural Networks with the back-propagation algorithm. Similarly to checkpointing techniques coming from the literature on Automatic Differentiation, it consists in dynamically selecting the forward activations that are saved (materialized) or deleted during the training process, and then automatically recomputing missing activations from those previously materialized. We propose an original computation model that combines two types of activation savings: either only storing the layer inputs, or recording the complete history of operations that produced the outputs. We prove that classical techniques from Automatic Differentiation literature do not apply in the fully heterogeneous case, since the classical assumption of memory persistence of materialized activations does not hold true anymore. We nevertheless provide an algorithm to compute the optimal computation sequence. 10:30 -11:00 / 14:30 – 15:00 Title: Running PINNs on the Grid5000: First Experiences, Romulo Silva, Debora Pina, Liliane Kunstmann, Daniel de Oliveira, Patrick Valduriez, Alvaro Coutinho, Marta Mattoso Abstract: The growing popularity of Neural Networks in scientific computing presents several challenges in configuring training parameters and validating the models. In mathematical problems, machine learning as a whole has been used to approximate computational costly problems, discover equations by coefficient estimation, and for building surrogates. Those applications are outside of the common usage of neural networks, and require a different set of techniques generally encompassed by Physics-Informed Neural Networks (PINNs), which appear to be a good alternative for solving forward and inverse problems governed by PDEs in a small data regime, specially when it comes to Uncertainty Quantification. PINNs have been successfully applied for solving problems in fluid dynamics, inference of hydraulic conductivity, velocity inversion, phase separation and many others, but we still need to investigate its computational aspects especially about its scalability when running in large-scale systems. Although there are hyperparameter setting recommendations, their fine-tuning is still required. In PINNs, this fine-tuning requires not only analyzing configurations but also how they relate to the loss function evaluation. We propose data provenance capture and analyses techniques to improve the model training of PINNs. We also report our first attempts to run PINNs in Grid5000. 11:00 -11:30 / 15:00 – 15:30 Title: Inferring phylogenetic networks from ancestral genes to regression trees and their use for trait evolution, Rafael de Souza Terra (afaelst@lncc.br) , Kary Ocana Abstract: Biological data provide detailed insights into the evolutionary history of species, traditionally represented as phylogenetic trees. Many evolutionary processes involve the transfer of genetic information, including vertical descent from parent to offspring, but also including so-called horizontal transfer, such as occurs via hybridization, recombination, introgression, gene transfer, and genome fusion. Networks are used for comparative analysis of continuous traits to describe various evolutionary events since all of these processes have presumably been occurring continuously throughout the history of life. So the phylogeny of life has been more or less network-like in different parts, depending on the balance between vertical and horizontal descent. Several open-source packages contribute to network estimation, manipulation and plotting, notably PhyloNet, BEAST2, TreeMix, Dendroscope, Galaxy platform, and PhyloNetworks. There are at least five heuristic uses of affinity networks in phylogenetics we considered at the modeling process: exploratory data analysis, displaying data patterns, displaying data conflicts, analyzing results, and testing phylogenetic hypotheses. Based on that idea, we have just used genes, trees, networks, and thought processes of phylogeneticists as a metaphor to model a network-based framework integrating python/R scripting libraries. We illustrate these results by classifying viruses with a classifier that we trained on simulated trees from different phylogeny networks. Next steps involve multiomic-data integration enriched with pathway networks to identify key transcriptional regulators and RNA binding proteins. 11:30-12:00 / 15:30 – 16:00 Break (Zoom break out room) 12:00 - 12:30 / 16:00 – 16:30 (Delayed) Title: Cache-based Workflow Scheduling - Daniel de Oliveira, Yuri Frota e Esther Abstract: Several today's scientific experiments are modeled as scientific workflows. Many of existing workflows share activities or sub-workflows. This way, it is interesting that a specific workflow execution could reuse data from previous executions. This reuse can be achieved when we have a cache-based execution where intermediate data can be gathered. There are some solutions to cache workflow data in an efficient way, such as the one proposed by Gaëtan Heidsieck et al. [2019]. The idea is to develop a scheduling heuristic that benefits from the cache mechanisms (i.e., the scheduling is cache-aware). The scheduling problem was modeled as a mixed integer optimization problem, which considers cached intermediate data to reuse while reducing execution time and financial cost of running workflow. Experiments performed with traces from Phenomenal workflow showed promising results. 12:00 - 12:30 / 16:00 - 16:30 Title: DJEnsemble : On the Selection of a Disjoint Ensemble of Deep Learning Black-Box Spatio-Temporal Models , Yania Souto, Rafael Pereira, Anderson Chaves, Rocio Zorrilla, Brian Tsan, Florin Rusu, Eduardo Ogasawara, Artur Ziviani, Fabio Porto Abstract: Consider a set of black-box models -- each of them independently trained on a different dataset -- answering the same predictive spatio-temporal query. Being built in isolation, each model traverses its own life-cycle until it is deployed to production. As such, these competitive models learn data patterns from different datasets and face independent hyper-parameter tuning. In order to answer the query, the set of black-box predictors has to be ensembled and allocated to the spatio-temporal query region. However, computing an optimal ensemble is a complex task that involves selecting the appropriate models and defining an effective allocation function that maps the models to the query region. In this presentation, we present a cost-based approach for the automatic selection and allocation of a disjoint ensemble of black-box predictors to answer predictive spatio-temporal queries. 13:00 - 13:15 / 17:17:15 - Closing